Intrinsically disordered proteins
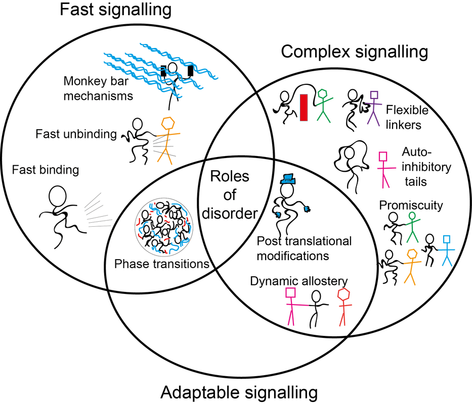
A significant proportion of eukaryotic proteins contain long disordered regions, which have no single stable structure and instead populate a myriad of highly heterogenous conformations that interchange very rapidly. These proteins are still functional within the cell. The particular over-representation of protein disorder within transcription factors suggests an important biological role for disorder within transcription factors. How functionally important is protein disorder for these critical regulatory proteins, why, and through what mechanism(s)?
The figure to the left summarises some potential mechanistic roles of protein disorder within transcriptional regulation. For more detailed discussion click here.
The figure to the left summarises some potential mechanistic roles of protein disorder within transcriptional regulation. For more detailed discussion click here.
CREB as an intrinsically disordered transcription factor
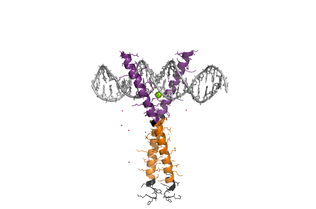
CREB is an extremely disordered transcription factor. It is a constitutively expressed member of the basic leucine zipper (bZIP) class of TFs, which is the largest and most conserved family of TFs that operate exclusively in eukaryotes, and also includes proto-oncogenes Fos and Jun. It is located downstream of key signalling and developmental pathways, and is thus involved with many cellular processes including cell differentiation, long-term memory formation, glucose metabolism and maintenance of the circadian rhythm. CREB overexpression is observed in bipolar disorder and Alzheimer’s Disease, as well as cancer. In contrast, CREB underexpression is associated with major depressive disorder.
bZIP TFs are grouped by their preferred DNA binding sites into 8 separate subfamilies. They are typically highly disordered, with several modules undergoing α-helical folding upon binding their targets (Figure 1). They bind to their DNA partners as dimers, with long, non-interrupted helices arranged in a “chopstick” fashion (see left). DNA binding is through the basic region (BR), each helix contacting a half-site of a cognate sequence on the DNA. Dimerization is controlled by a leucine zipper (LZ) region; two parallel coiled-coil amphipathic α-helices wrapped around each other. Finally, the transactivation domains, which bind to co-activator proteins to initiate assembly of the transcriptional machinery, can also fold into α-helices. CREB has two activation domains; Q2 can act alone to stimulate low levels of transcription, but transcription is increased if the kinase inducible domain (KID) binds the transcriptional co-activator CBP. CREB is one part of an enormous complex, that results in transient recruitment of RNApolII and transcription.
We are investigating what functional mechanistic roles protein disorder might play in transcriptional regulation by CREB.
bZIP TFs are grouped by their preferred DNA binding sites into 8 separate subfamilies. They are typically highly disordered, with several modules undergoing α-helical folding upon binding their targets (Figure 1). They bind to their DNA partners as dimers, with long, non-interrupted helices arranged in a “chopstick” fashion (see left). DNA binding is through the basic region (BR), each helix contacting a half-site of a cognate sequence on the DNA. Dimerization is controlled by a leucine zipper (LZ) region; two parallel coiled-coil amphipathic α-helices wrapped around each other. Finally, the transactivation domains, which bind to co-activator proteins to initiate assembly of the transcriptional machinery, can also fold into α-helices. CREB has two activation domains; Q2 can act alone to stimulate low levels of transcription, but transcription is increased if the kinase inducible domain (KID) binds the transcriptional co-activator CBP. CREB is one part of an enormous complex, that results in transient recruitment of RNApolII and transcription.
We are investigating what functional mechanistic roles protein disorder might play in transcriptional regulation by CREB.